MGI and Sentieon jointly release DNAscope MGI workflow performance…
At the ASHG conference held in October 2022, MGI and Sentieon jointly released performance data for the optimized DNAscope workflow for DNBSEQ-G400. They also announced plans to engage in close collaboration to promote the clinical and research applications of whole-genome sequencing, serving users in North America and other regions.

MGI DNBSEQ-G400 is a desktop-level, mid-throughput sequencing platform with a new sequencing slot system and optimized optical and biochemical systems, providing users, especially clinical and research users, with a highly flexible and low-cost sequencing platform. “This system can produce up to 1,440GB of data per day and supports flexible selection of multiple sequencing slots with different throughputs, generating read lengths from SE50 to PE300. The DNBSEQ-G400 was released in the US market in August of this year and was deployed to multiple customer labs within a week,” said Dr. Yongwei Zhang, CEO of MGI Americas. “Accuracy and cost advantage are the core strengths that MGI can provide to local customers, especially when combined with the Sentieon analysis workflow. The entire system can provide more accurate clinical mutation detection results, helping users develop more precise diagnostic and treatment plans.”

“Sentieon is very pleased to provide optimized secondary analysis solutions for MGI and to jointly release this solution with MGI at ASHG,” said Dr. Jun Ye, CEO of Sentieon. “This optimized analysis workflow will help users of the MGI platform to perform mutation detection analysis more efficiently, quickly, and accurately. We look forward to continuing to work closely together to provide higher quality solutions to the genomics industry.”
Sentieon’s DNAscope machine learning analysis workflow can match different sequencing platforms with different model files and generate various data types from library capture kits. In a recently published methodological preprint, the data showed that DNAscope’s whole-genome and exome accuracy far exceeded standard open-source analysis workflows.

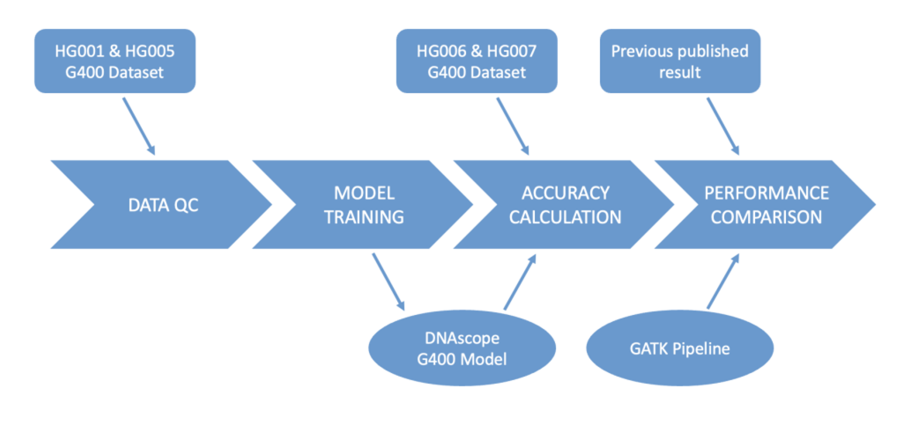
In this collaboration, teams from Sentieon and MGI analyzed the standard sample data of MGI DNBSEQ-G400 to obtain various data quality control indicators such as GC bias and quality value distribution. Based on this, the DNAscope process was optimized and adjusted, and the MGI v0.5 model file was trained using HG001 and HG005 data, followed by accuracy calculation on the HG006 and HG007 test datasets. Specifically, the FASTQ files of the HG001 and HG005 datasets (including PE100 and PE150 read lengths) were aligned to the hg38 reference genome using Sentieon BWA, and after quality control, BAM files were downsampled to generate multiple training sets with sequencing depth coverage of 15x-45x. HG001-PE150-30x data was excluded from the training set as hold-out. Comparing the candidate mutation sites generated with the true set VCF sites of GIAB v4.2.1, the DNAscope model training tool generated a gradient boosting decision tree model file named MGI v0.5 model.
The MGI v0.5 model was proven to be highly accurate in subsequent testing. We evaluated its accuracy using the standard sample data of HG006 and HG007 generated by the G400 sequencer, as well as the HG001-PE150-30x data that was set aside separately, and calculated false positives, false negatives, and overall accuracy F1-Score (using hap.py 0.3.10 and RTGtools vcfeval 3.9.2). For reference, the results shown in the table below include the performance of the old version of the MGI model published in 2019, as well as the corresponding reference dataset of Illumina.
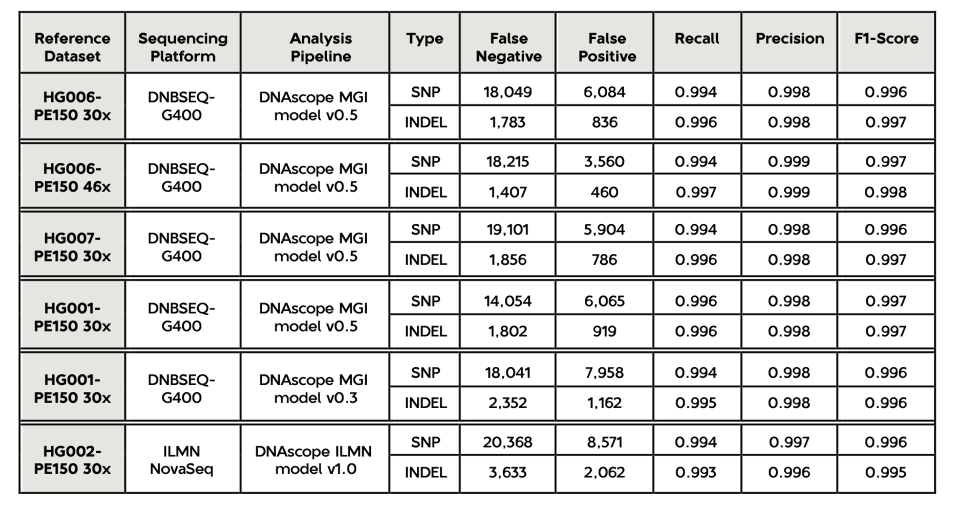
From the results, we can observe the following:
- Compared with Illumina NovaSeq standard data at the same depth (data from Sentieon DNAscope white paper preprint, which introduces machine learning methods and the new model’s performance), the accuracy of DNBSEQ-G400 + DNAscope is significantly higher, especially in terms of Indel errors, which are reduced to only half of Illumina’s (~2.7k vs. 5.6k). It should be noted that there may be multiple reasons for the difference in accuracy due to different standard data.
- Compared with the old data (MGISEQ-2000) + old model (MGI model v0.3) published in 2019, the current version’s accuracy has been further improved, reducing 20%-25% of detection errors. Considering the decrease in the cost of the current version’s reagents and the improvement in process computing speed, the overall performance improvement is more apparent.
- HG006 46x depth data (the normal output data of one lane of DNBSEQ-G400) exhibits higher accuracy than its 30x depth data, proving that data depth above 30x still contributes to accuracy.
- The accuracy of the HG001 training set left out is very close to the HG006 and HG007 test sets, indicating that the model did not overfit the training set.
In this collaboration, the teams on both sides optimized the DNAscope process for DNBSEQ-G400’s new data and trained a dedicated adapter model for MGI v0.5. The PCR-free whole-genome sequencing of DNBSEQ-G400 in library construction and sequencing avoids PCR amplification, greatly reducing the error rate. Meanwhile, DNAscope avoids errors in the variation detection process by specifically identifying the error patterns of the MGI sequencing platform through machine learning methods. In the accuracy evaluation, this set of sequencing analysis processes has significantly improved compared to previously published results and exceeded the current mainstream sequencing analysis solutions in the industry.
To make it more convenient for users to use this integrated solution, MGI announced at ASHG that it would provide Sentieon licenses free of charge to some users in North America to form a variation detection solution with the sequencing instrument.