DNAscope LongReads SV – a structural variation detection tool…
In recent years, third-generation sequencing platforms represented by PacBio and ONT have been flourishing. ONT released Q20+ reagents, which brought the accuracy of reads close to that of second-generation sequencing, while PacBio announced at the 2022 ASHG that the cost of whole-genome sequencing on their new platform has been reduced to $1000, providing a cost-effective solution for the clinical and research applications of third-generation sequencing. The Sentieon bioinformatics software development team has been working closely with industry partners to develop Sentieon Minimap2, an acceleration alignment module for PacBio and ONT data, and a short variant detection workflow for SNVs/Indels
We know that the biggest advantage of third-generation data is that it can better detect large-scale structural variations through long read lengths. However, current mainstream variant detection software in the industry has room for improvement in terms of accuracy and speed. The Sentieon team has developed DNAscope LongReads SV, a structural variant detection tool based on haplotype phasing, which is highly efficient and accurate in detecting structural variations and can achieve single-base resolution, specifically designed for ONT and PacBio data.
We evaluated the accuracy of SV detection using a dataset from both ONT and PacBio platforms, including r9 and new Q20 reagents for ONT, and NIST SV Truth v0.6 from standard sample HG002 for evaluation regions. We selected the popular Sniffles2 software as the comparison workflow, and also included the popular PbSV in the comparison for PacBio data evaluation. SV accuracy was calculated using Hap-eval, a recently released open-source tool for structural variation accuracy evaluation developed by Sentieon for their GMKF third-generation sequencing analysis project.
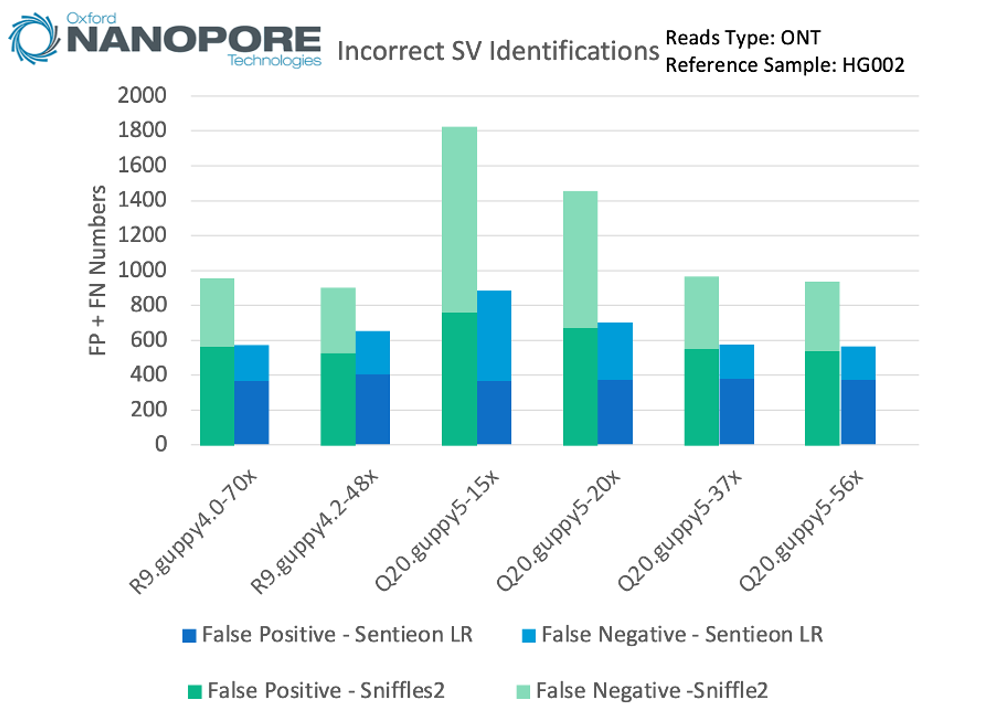
Using ONT data to analyze the whole genome, it can be seen that the accuracy of Sentieon LongReads SV exceeds Sniffles2 on all datasets, with significantly reduced false positives and false negatives. The updates of sequencing reagents and base callers have contributed to the accuracy of SV, but not significantly. In addition, the Sentieon workflow is particularly suitable for low-depth data. Even at a depth of 15x, the Sentieon workflow can still control the total number of errors within 1000 in the whole genome.
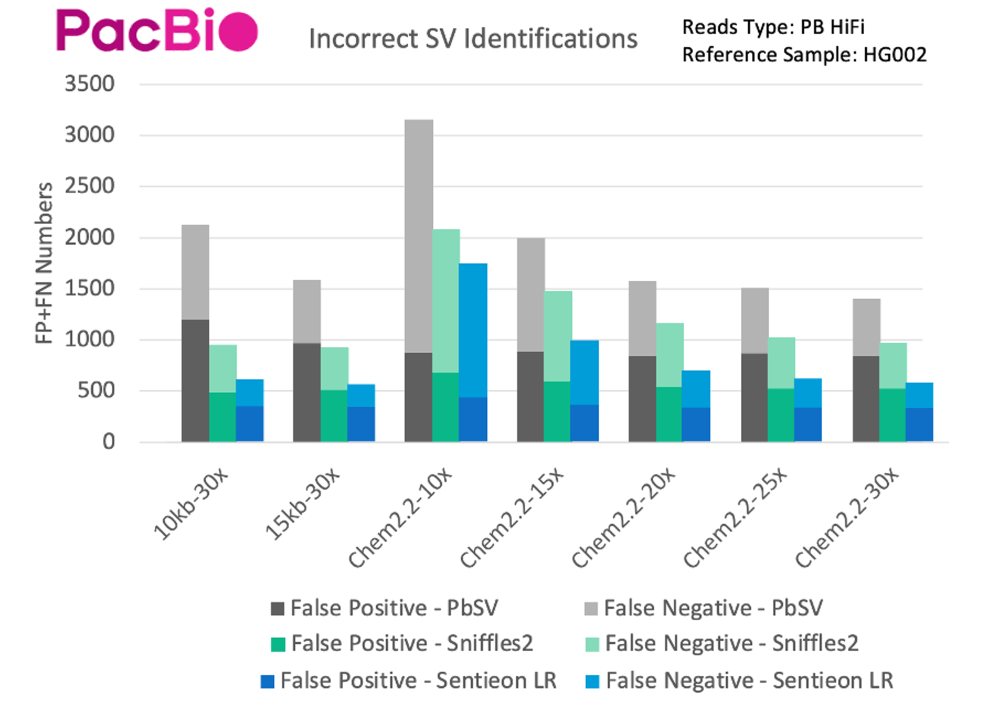
The situation is similar for PacBio data, with the highest accuracy achieved by the Sentieon pipeline, regardless of read length or sequencing depth, and a relatively large improvement in accuracy compared to Sniffles2 and PbSV. Two conclusions that can be drawn are that at the same depth, ONT’s SV detection is slightly better than PacBio HiFi, and higher depth does not contribute much to SV detection when sequencing depth exceeds 20x.
In addition to structural variations, long-read data can also be used for SNP and Indel detection on the basis of phasing, and can also touch on regions that cannot be aligned by second-generation sequencing, such as important clinical genes like SMN1. Illumina sequences can hardly cover the exon regions of SMN1, while third-generation sequencing can cover the entire gene more evenly and detect accurate disease-causing variations.
With the accelerated version of Minimap2 and integrated SV, SNP/Indel detection pipelines, the Sentieon software can effectively support the development of clinical analysis workflows for third-generation sequencing users.
The Sentieon team will continue to collaborate with partners in the third-generation sequencing ecosystem to jointly develop and validate new data analysis workflows, pushing long-read data analysis into more applications such as cancer detection, methylation analysis, and contributing to the scientific research and clinical use in the industry.