PrecisionFDA Multi-Omics AI Challenge: Sentieon Wins Again – Biomarker…
Recently, PrecisionFDA announced the results of its second Multi-Omics Joint Analysis AI Challenge, in which the Sentieon team once again claimed the overall championship and won all sub-item championships. Sentieon’s two AI models ranked in the top two among all 22 participating models. Please click “Read More” at the end of the article to view the PrecisionFDA results announcement page.
The full name of this challenge is the PrecisionFDA Brain Cancer Predictive Modeling and Biomarker Discovery Challenge, which lasted for three and a half months from November 2019 to mid-February 2020. It was divided into three sub-item challenges and attracted dozens of teams from around the world, including large pharmaceutical companies, research and medical institutions, and renowned universities. In each sub-item competition, participating teams needed to design and train AI models using different omics data, automatically discover relevant biomarkers in samples, and predict the final clinical treatment outcomes for patients.
The organizers specifically introduced the championship team: Sentieon’s solution selected 46 biomarkers, including the expression levels of 40 genes, 4 copy variation regions, and 2 phenotype features.
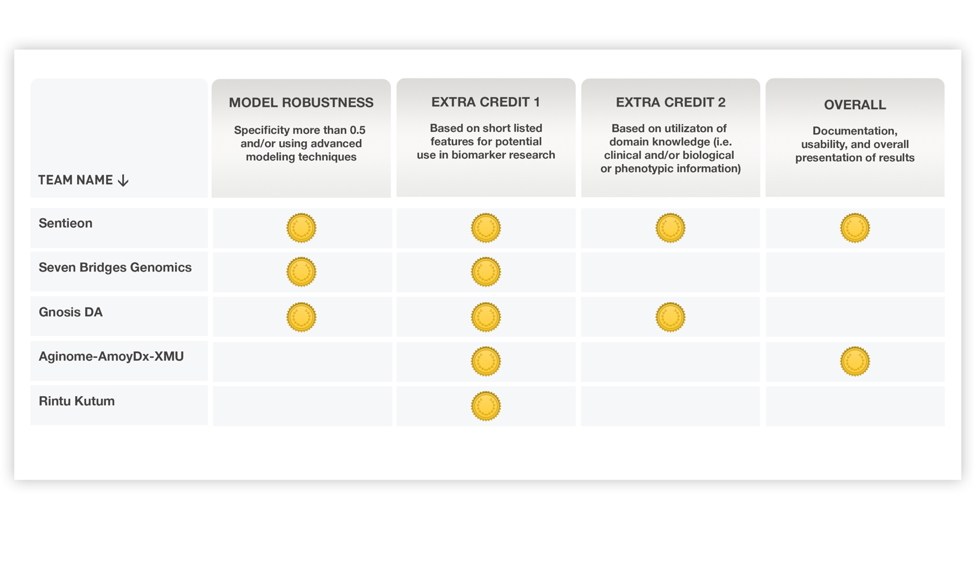
In the final evaluation, the accuracy of Sentieon’s team model and the additional scoring features of the modeling process methodology both received the “Medal” award from the organizer.
Previous Results
This is Sentieon’s fifth time participating in the precisionFDA challenge. As a team focused on bioinformatics analysis algorithms, Sentieon has won multiple championships in every challenge.

Challenge Background
In September 2018, as a cutting-edge exploration platform in the field of precision medicine, precisionFDA organized the first medical multi-omics data challenge, inviting submissions from around the world to provide solutions. Participating teams needed to conduct joint analysis on gene expression profiles, proteomics, and clinical phenotype data, use machine learning to model the data, and find and correct incorrectly labeled sample tags. The detailed background and specific settings of the challenge, as well as information on Sentieon’s championship results, have been described in previous reports and will not be repeated here. For more details, please click on “RECOMB News: Sentieon Wins precisionFDA Multi-Omics Data Challenge.”
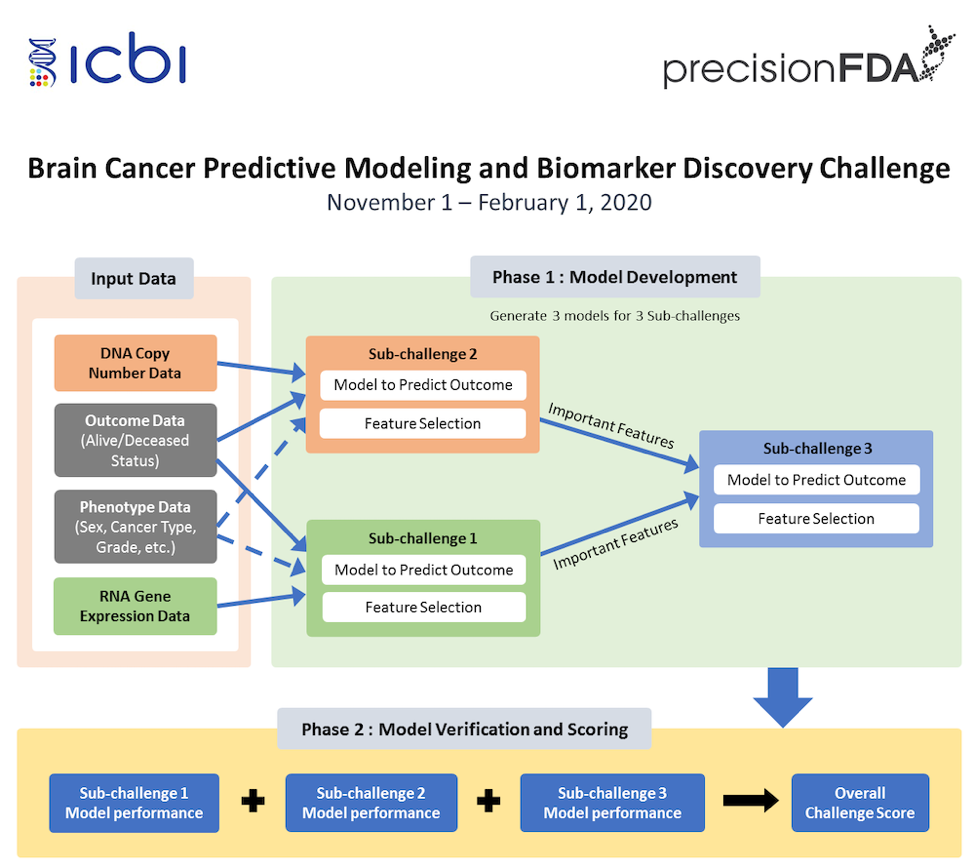
As a further exploration, precisionFDA launched a new multi-omics joint analysis challenge in November 2019, in conjunction with clinical data from Georgetown University, inviting participating teams to establish machine learning AI models based on brain tumor patients’ gene expression profiles, genomic copy number variations (CNV), and phenotype data to predict the patients’ clinical treatment outcomes (outcome data).

Like previous precisionFDA challenges, this challenge attracted dozens of well-known institutions from around the world, including Seven Bridges, Roche, AGS, MSKCC, Aginome, Garvan, and FDA, representing the leading edge of the industry in this field.
Challenge Setup
Like previous precisionFDA challenges, this challenge attracted dozens of well-known institutions from around the world, including Seven Bridges, Roche, AGS, MSKCC, Aginome, Garvan, FDA, etc., representing the industry’s leading level in this field.
Competition Setting
The challenge was divided into two stages: In the first stage, all data of the training samples were provided for participating teams to design and train models. In the second stage, new test sample data was provided, which only included the patient’s gene expression profile, genome copy number variation, and phenotype data. The participating teams were required to predict the patient’s clinical treatment effect based on the model established in the first stage. The second stage of the competition was divided into three sub-challenges according to the input data type: Sub-Challenge 1 could only use gene expression profiles and phenotype data for prediction, Sub-Challenge 2 could only use genome copy number variation and phenotype data, while Sub-Challenge 3 required the comprehensive analysis of these three types of data. Finally, the organizers compared the actual clinical treatment effect with the submitted prediction effect, and comprehensively considered the accuracy, sensitivity, and specificity of the prediction results. The final comprehensive ranking was the weighted sum of the rankings of the three sub-items.
It can be seen that in the competition, multi-omics data is a typical “high-dimensional” problem. There are more than 20,000 gene expression levels, genome copy number variations, and a large amount of phenotype data that need to be matched with a very small number of samples, testing the team’s methods for automatically finding effective features. Other challenges in the data include the very imbalanced distribution of phenotype data and clinical treatment effect data, and the complexity of genome copy number variation data types, which require careful preprocessing before modeling. At the same time, the clinical treatment effect of tumors itself is also a problem with many influencing factors, but its accurate prediction is of great significance for selecting and evaluating treatment plans, as well as drug development.
Results announcement
In the end, after the 3.5-month competition, the Sentieon team submitted two models that respectively achieved excellent results of first and second place in total score (the lower the “score” value, the better).
| Rank | Team | Sub-challenge 1 | Sub-challenge 2 | Sub-challenge 3 | Overall Score |
| 1 | Sentieon | 19 | 8 | 5 | 37 |
| 1 | Sentieon | 12 | 12 | 17 | 58 |
| 2 | Seven Bridges Genomics | 49 | 18 | 12 | 91 |
| 3 | Gnosis DA | 24 | 34 | 20 | 98 |
| 4 | Aginome-AmoyDx-XMU | 28 | 24 | 25 | 102 |
| 5 | Rintu Kutum | 31 | 36 | 18 | 103 |
Due to its excellent performance in methodology, Sentieon was awarded all four additional “medals” by the organizers.
The organizers introduced the winning Sentieon model, which selected a total of 46 features, including 40 gene expressions, 4 copy variation regions, and 2 phenotype features, all of which are biomarkers related to brain tumor progression and play important roles in clinical diagnosis and drug development.
Similar to the first multi-omics challenge, Sentieon, as the winning team of this challenge, will share detailed modeling methods through speeches and published articles. We will continue to report on this in the form of WeChat articles.
Significance of the Challenge
In recent years, with the vigorous development of big data projects in the field of biomedical research, the industry generally believes that artificial intelligence can be applied in various directions. However, the currently popular applications are still limited to assisting image interpretation or assisting diagnosis based on natural language processing, and there are few mature applications in multi-omics joint analysis. One reason for this is that the industry’s various technical routes are mixed, and there is a lack of a credible platform to evaluate and discuss different methods. We are pleased to see that precisionFDA has organized such challenges in a timely manner to recommend excellent methods and teams to the industry!
Another limitation to the application of multi-omics joint analysis is the scarcity of training sets. Building a true multi-omics dataset requires collecting multiple data including genomics, transcriptomics, proteomics, genomic copy number variations, metabolomics, methylation, and phenotypic data, which is very costly and requires a large amount of manpower to maintain. A scarce training set means that general deep learning methods are difficult to directly transfer to the analysis of multi-omics data. Sentieon demonstrated excellent algorithm development and practical capabilities in this challenge, achieving high-dimensional modeling on a very small sample set. This is particularly important in the establishment of medical multi-omics models and lays a solid technical foundation for the development of subsequent practical applications.
The Sentieon team has long been committed to the development of core algorithms for bioinformatics software and medical big data AI models. We are very willing to cooperate with clinical diagnosis companies and drug development enterprises in the industry to explore data processing solutions and help users more accurately use multi-omics data to solve clinical problems!