Embracing CPU Acceleration Solution – AWS and Sentieon Jointly…
Recently, AWS and Sentieon jointly released an evaluation report that deployed the Sentieon acceleration analysis workflow on AWS’s latest Hpc6a high-performance computing instance, demonstrating the triple advantages of computing cost, analysis speed, and result accuracy.
Click to access https://aws.amazon.com/blogs/hpc/cost-effective-and-accurate-genomics-analysis-with-sentieon-on-aws/

The beginning of 2023 has been exciting for customers in the genomics industry, with sequencing platforms such as Illumina/MGI/Element/Ultima announcing sequencing costs below $2 per GB of data. PacBio also announced through its Revio platform to lower the cost of third-generation sequencing to the range of $10 per GB of data.
While these newly released platforms offer new options for genome sequencing, each platform has its own specific data features and error paradigms. Considering that sequencing data may increase significantly, diverse sequencing schemes also pose new challenges for secondary data analysis. Customers urgently need more efficient and accurate solutions for processing genomic data, and we need to be flexible enough to handle data generated by all sequencing platforms.
Sentieon has developed a series of high-performance tools for processing and analyzing genomic data, providing fast and accurate industrial-grade software solutions for secondary analysis of NGS. Among them, the DNAseq workflow provides a 10x acceleration compared to the classical GATK best practice workflow, while DNAscope applies machine learning algorithms to improve accuracy and adapt to various sequencing platforms on top of accelerated analysis.
In this article, the authors tested the performance of Sentieon’s DNAseq and DNAscope workflows using publicly available datasets from Illumina, PacBio HiFi, Element Biosciences, and Ultima Genomics platforms on Amazon Elastic Compute Cloud (Amazon EC2) instances. Readers will learn about the runtime, computational cost, and accuracy of running whole-genome data secondary analysis in various AWS instances. The download link for the evaluation data, the selection of AWS computing environment, and the calculation of result accuracy are all explained in detail in the original blog post.
Performance of Sentieon workflow on Hpc6a instance
Sentieon software adopts a CPU acceleration scheme that can be flexibly deployed on various EC2 instance types. The recently released Hpc6a instance by AWS provides extremely high cost-effectiveness for compute-intensive tasks, making it particularly suitable for Sentieon’s secondary analysis workflow. The figure below shows the analysis runtime and on-demand compute prices when running cross-genome sequencing platform analysis on the hpc6a.48xlarge instance in the US East region.
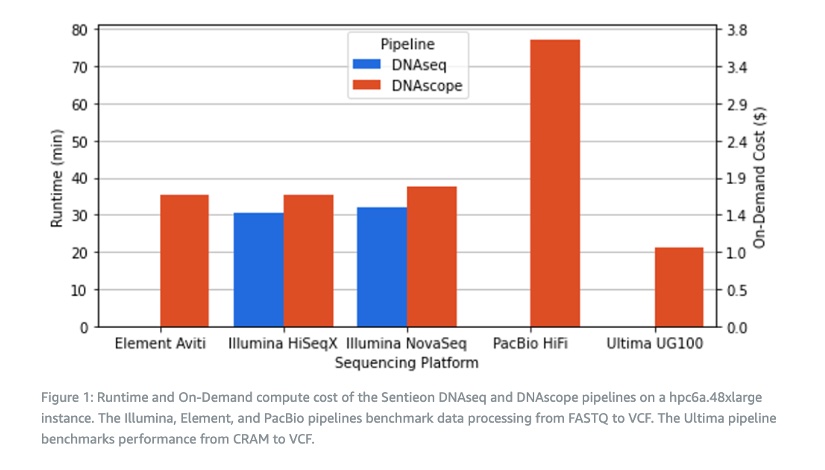
It can be seen that on the hpc6a.48xlarge instance, the Sentieon DNAseq process took 32 minutes to analyze 30x Illumina NovaSeq data from FASTQ to VCF. Compared to DNAseq, the DNAscope process reduced SNP errors by 53% and INDEL errors by 78% on the same data while only taking an additional 3-5 minutes. The on-demand computing cost for both DNAseq and DNAscope processes is usually around 1.5-1.8 USD, equivalent to about 10-12 CNY.
It’s worth noting that the DNAscope LongRead process, which handles PacBio HiFi data, has a larger computational load compared to short-read data as it involves multiple rounds of variant calling and phasing. This process was completed in 77 minutes with an on-demand cost of 3.7 USD. The Element Biosciences AVITI system is a new desktop sequencer that was launched in the spring of 2022 and is supported by an optimized Sentieon DNAscope process. This process was completed in 35 minutes with an on-demand cost of 1.7 USD. Finally, the starting data for the Ultima UG100 has already been aligned, so we only performed variant calling. The CRAM to VCF conversion was completed in 22 minutes with an on-demand cost of 1.1 USD on the hpc6a.48xlarge instance.
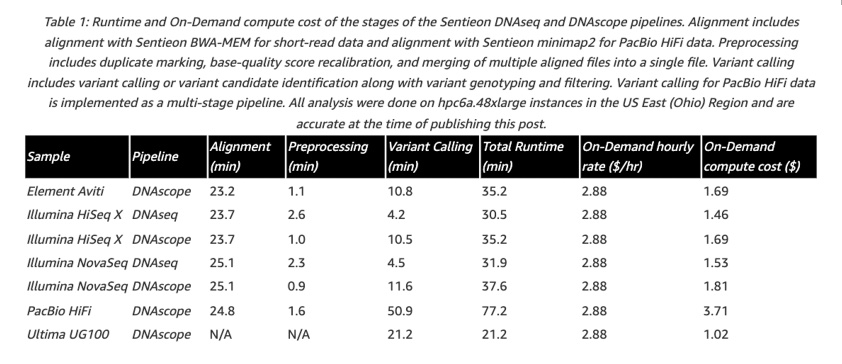
The following figure shows detailed data on computing time and costs for each step.
Benchmarks on more AWS instances
Sentieon software has high scalability and can use large instances to speed up single-sample analysis or small instances to process small samples such as panels. To explore the cost range of Sentieon software, we conducted benchmark tests of the Sentieon DNAseq process using Illumina NovaSeq datasets on various Amazon EC2 instance types. These tests include x86 architecture represented by Intel and AMD, and ARM architecture represented by Graviton. The running speed, on-demand, and spot instance computing costs of the Sentieon DNAseq process (from FASTQ to VCF) on these instances are shown in the following figure. To achieve the fastest analysis speed, the DNAseq process can complete a 30x whole-genome on a c6a.48xlarge instance in 24 minutes, with an on-demand cost of 2.9 US dollars. In addition to the hpc6a.48xlarge mentioned earlier, the c7g.8xlarge instance also provides good computing costs, with an on-demand price of 2.3 US dollars.
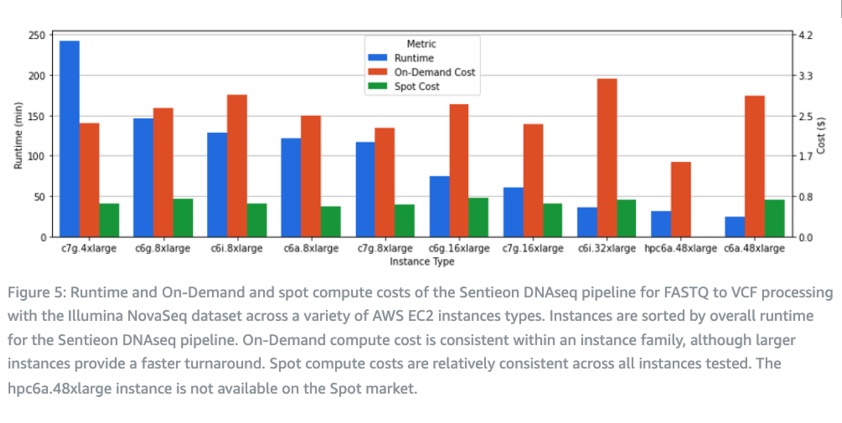
These results highlight the high utilization of computing resources by Sentieon software, which can adapt to both small and large instances. It is worth noting that in this evaluation, we only included compute-optimized instances, but Sentieon tools can also be used with other EC2 instance families.
DNAseq and DNAscope Pipeline Accuracy Demonstration
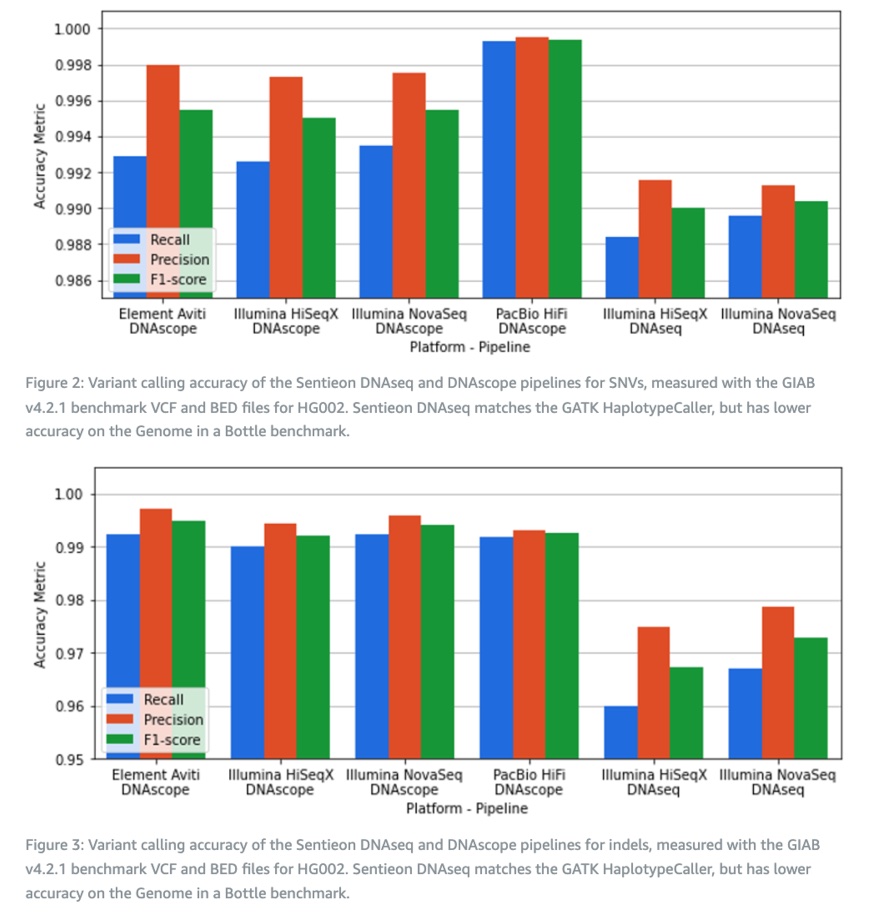
The authors calculated the variation detection accuracy of DNAseq and DNAscope processes based on the HG002 reference standard and GIAB v4.2.1 truth set. Similar to previously published results, the machine learning-based DNAscope process can provide highly accurate SNP and Indel detection on all sequencing platforms, with F1 scores for SNP and Indel detection exceeding 99.5% and 99.2%, respectively. The DNAseq process provides the same results as the GATK best practices process, but with lower accuracy compared to DNAscope. The Indel accuracy of PacBio HiFi data is slightly lower, but the SNP accuracy exceeds that of all short-read data. Finally, in the analysis results of DNAscope, the SNP accuracy of Ultima UG100 reached the benchmark of other short-read platforms.
Summary
Sentieon’s DNAscope workflow provides accurate and rapid secondary analysis on various sequencing platforms, which can be deployed on various EC2 instances on AWS by adapting machine learning models. These workflows are highly scalable, ranging from 192 vCPU c6a.48xlarge instances for single-sample analysis in less than 24 minutes, to c7g.4xlarge instances for more flexible use of spot prices.
AWS’s Hpc6a instances provide highly competitive computing power and cost, with hpc6a.48xlarge instances supporting Sentieon workflows that can process a 30x whole genome in 32 minutes at an on-demand cost of $1.5. In addition, the on-demand analysis costs for most other c6i, c6a, c6g, and c7g instances are below $3 for whole genome analysis.